Abstract: In this paper, we question if self-supervised learning provides new properties to Vision Transformer (ViT) [18] that stand out compared to convolutional networks (convnets). Beyond the fact that adapting self-supervised methods to this architecture works particularly well, we make the following observations: first, self-supervised ViT features contain explicit information about the semantic segmentation of an image, which does not emerge as clearly with supervised ViTs, nor with convnets. Second, these features are also excellent k-NN classifiers, reaching 78.3% top-1 on ImageNet with a small ViT. Our study also underlines the importance of momentum encoder [31], multi-crop training [10], and the use of small patches with ViTs. We implement our findings into a simple self-supervised method, called DINO, which we interpret as a form of self-distillation with no labels. We show the synergy between DINO and ViTs by achieving 80.1% top-1 on ImageNet in linear evaluation with ViT-Base.
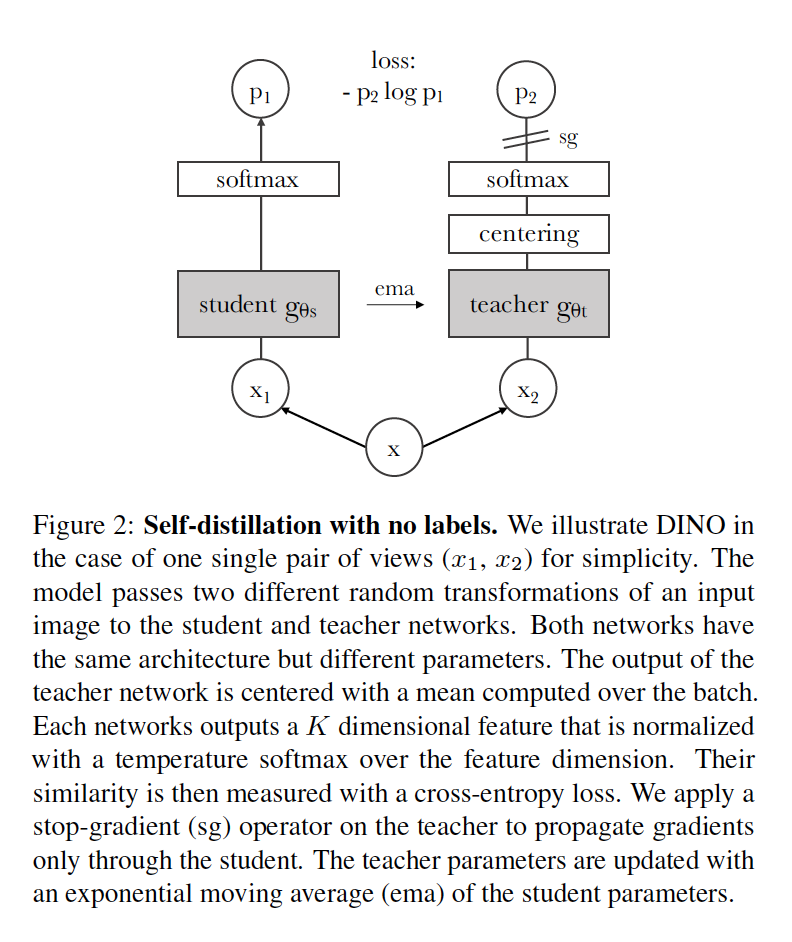
Own Summary: In this paper authors show effectiveness of the combination of DINO framework and ViT based architectures such as ViT and DEIT. There is no contrastive training nor negative pairs, rather ideas such as momentum encoder and multi-crop augmention from BYOL and SWAV respectively are adapted. They use distillation with a teacher-student configuration, and avoid representation collapse by centering and sharpening target distributions generated by the teacher. 2 large views (~50%) are used as targets and all views (2 large, 4 small) are used for predictions similar to SWAV. Centering values and teacher parameters are updated via ema (exponential moving average).
aug_pipelines = get_dino_aug_pipelines()
bs = 4
x_large = [torch.randn(4,3,224,224)]*2
x_small = [torch.randn(4,3,96,96)]*4
deits16 = deit_small(patch_size=16, drop_path_rate=0.1)
deits16 = MultiCropWrapper(deits16)
dino_head = DINOHead(deits16.encoder.embed_dim, 2**16, norm_last_layer=True)
student_model = nn.Sequential(deits16,dino_head)
deits16 = deit_small(patch_size=16)
deits16 = MultiCropWrapper(deits16)
dino_head = DINOHead(deits16.encoder.embed_dim, 2**16, norm_last_layer=True)
teacher_model = nn.Sequential(deits16,dino_head)
dino_model = DINOModel(student_model, teacher_model)
dino_model.student[1]
Training schedule for DINO
fig,ax = plt.subplots(1,2,figsize=(15,5))
lr_sched = combine_scheds([0.1,0.9], [SchedLin(0.,1e-3), SchedCos(1e-3,1e-6)])
ax[0].plot([lr_sched(i) for i in np.linspace(0,1,100)]);ax[0].set_title('lr')
wd_sched = SchedCos(0.04,0.4)
ax[1].plot([wd_sched(i) for i in np.linspace(0,1,100)]);ax[1].set_title('wd');
